The globe artichoke genome is now available for use and four globe artichoke genotypes have been recently re-sequenced.
- D. Scaglione, S. Reyes-Chin-Wo, A. Acquadro, L. Froenicke, E. Portis, C. Beitel, M. Tirone, R. Mauro, A. Lo Monaco, G. Mauromicale, P. Faccioli, L. Cattivelli, L. Rieseberg, R. Michelmore & S Lanteri. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 6, 19427; doi: 10.1038/srep19427 (2016)
- A. Acquadro, L. Barchi, E. Portis, G. Mangino, D. Valentino, G. Mauromicale & S Lanteri. Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation. Sci.Rep. 7; 5617; doi:10.1038/s41598-017-05085-7 (2017)
- A. Acquadro, E. Portis, D. Valentino, L. Barchi & Sergio Lanteri. “Mind the Gap”: Hi-C Technology Boosts Contiguity of the Globe Artichoke Genome in Low-Recombination Regions. G3: Genes, Genomes, Genetics 10, 3557-3564; doi:10.1534/g3.120.401446 (2020)
It represents the first published genome sequence of a Compositae crop species fully available to the scientific community.
The genome sequence has been unravelled by a consortium including the University of Torino (DISAFA, Plant Genetics and Breeding, Italy, team leader Sergio Lanteri), the University of California (The Genome Center, Davis, CA, USA, team leader Richard Michelmore) and the Università di Catania (Di3A, Italy, team leader Giovanni Mauromicale) in the framework of the Compositae Genome Project (CGP).
The project started in 2011 and later on was joined by the University of British Columbia (Canada, team leader Loren Rieseberg) and Crea (Genomics Research Centre, Fiorenzuola d’Arda, Italy, team leader Luigi Cattivelli).
The genome draft was assembled from ~133-fold next-generation sequencing data, into 13K scaffolds (N50= 125 Kbp, L50=1411), which represent 725 Mb of genomic sequence with a de novo prediction of 26,906 gene models. Thanks to a low coverage whole genome-sequencing of an F1 highly segregating mapping population, a dense genetic map based on the two-way pseudo test cross strategy, was built up and used for anchoring and orienting 73% of the assembled scaffolds in 17 reconstructed pseudomolecules.
This was achieved thanks to the development of a novel pipeline, namely SOILoCo (Scaffold Ordering by Imputation with Low Coverage) https://bitbucket.org/dscaglione-igatech/soiloco.
In 2017, the resequencing analyses (~35X) of four globe artichoke genotypes, representative of the core varietal types, as well as a genotype of the related taxa cultivated cardoon was carried out. The genomes were reconstructed at a chromosomal scale and structurally/functionally annotated. Gene prediction indicated a similar number of genes, while distinctive variations in miRNAs and resistance gene analogues (RGAs) were detected. Overall, 23,5M SNP/indel were discovered (range 6,34M -14,50M). The impact of some missense SNPs on the biological functions of genes involved in the biosynthesis of phenylpropanoid and sesquiterpene lactone secondary metabolites was predicted. The identified variants contribute to infer on globe artichoke domestication of the different varietal types, and represent key tools for dissecting the path from sequence variation to phenotype. The new genomic sequences are fully searchable through independent Jbrowse interfaces, which allow the analysis of collinearity and the discovery of genomic variants, thus representing a one-stop resource for C. cardunculus genomics
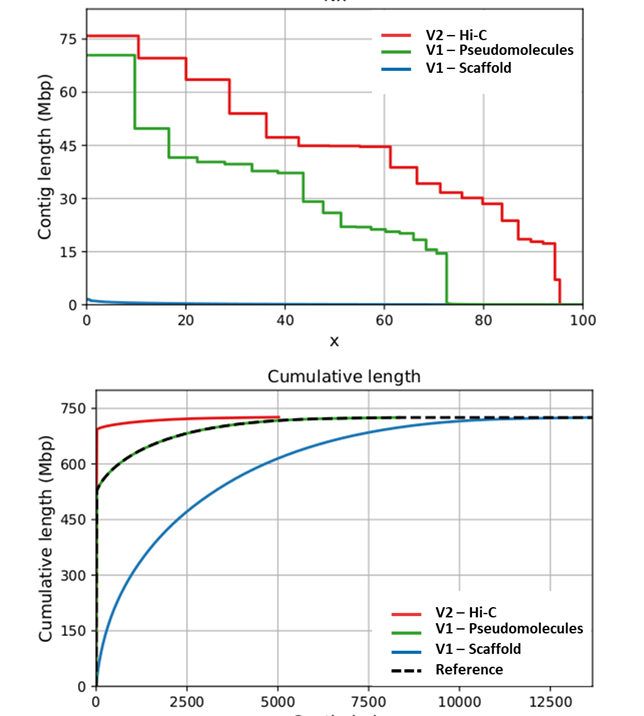
Recently, based on v.1.0 sequencing data, we generated a new genome assembly (v2.0), obtained from a Hi-C (Dovetail™) genomic library, and which improves the scaffold N50 from 126 kb to 44.8 Mb (~356-fold increase) and N90 from 29 kb to 17.8 Mb (~685-fold increase). While the L90 of the v1.0 sequence included 6,123 scaffolds, the new v2.0 just 15 super-scaffolds, a number close to the haploid chromosome number of the species. The cumulative size of unplaced scaffolds in v.2.0 was reduced of 165 Mb, increasing to 94% the anchored genome sequence. The marked improvement is mainly attributable to the ability of the proximity ligation-based approach to deal with both heterochromatic (e.g.: peri-centromeric) and euchromatic regions during the assembly procedure, which allowed to physically locate low recombination regions. The new high-quality reference genome enhances the taxonomic breadth of the data available for comparative plant genomics and led to a new accurate gene prediction (28,632 genes), thus promoting the map-based cloning of economically important genes.
(C. cardunculus var. scolymus, genotype ‘2C’’)
- v2.0 – Hi-C Superscaffolds
Annotation (Acquadro et al. 2020) on v2.0 assembly
- v2.0 – Genomic features
- v2.0 – Transcripts
- v2.0 – Proteins
(C. cardunculus var. scolymus, genotype ‘2C’)
- v1.0 – Pseudomolecules
- v1.0 – Scaffolds
- v1.0 – Un-anchored scaffolds
v1.0 – annotation (Scaglione et al 2016) on v1.0 assembly
- v1.0 – Genomic features
- v1.0 – Transcripts
- v1.0 – Proteins
v1.1 – annotation (Acquadro et al 2017) on v1.0 assembly
- v1.1 – Genomic features
- v1.1 – Transcripts
- v1.1 – Proteins
zip file (genome & features)
- A41 (Cultivated Cardoon)
- Spinoso di Palermo (‘Spinosi’ type)
- Violetto di Sicilia (‘Catanesi’ type)
- Violetto di Toscana ( ‘Violetti’ type)
- Romanesco C3 (‘Romaneschi’ type)

